Love Python and seismic? You need xarray
/If you use Python on a regular basis, you might have noticed that Pandas is eating everything, not just things made out of bamboo.
But one thing Pandas is not useful for is 2-D or 3-D (or more-D) seismic data. Its DataFrames are implicitly two-dimensional tables, and shine when the columns represent all sorts of different quantities, e.g. different well logs. But, the really powerful thing about a DataFrame is its index. Instead of having row 0, row 1, etc, we can use an index that makes sense for the data. For example, we can use depth for the index and have row 13.1400 and row 13.2924, etc. Much easier than trying to figure out which index goes with 13.1400 metres!
Seismic data, on the other hand, does not fit comfortably in a table. But it would certainly be convenient to have real-world coordinates for the data as well as NumPy indices — maybe inline and crossline numbers, or (UTMx, UTMy) position, or the actual time position of the timeslices in milliseconds. With xarray we can.
In essence, xarray brings you Pandas-like indexing for n-dimensional arrays. Let’s see why this is useful.
Make an xarray.DataArray
The DataArray object is analogous to a NumPy array, but with the Pandas-like indexing added to each dimension via a property called coords. Furthermore, each dimension has a name.
Imagine we have a seismic volume called seismic with shape (inlines, xlines, time) and the inlines and crosslines are both numbered like 1000, 1001, 1002, etc. The time samples are 4 ms apart. All we need is an array of numbers representing inlines, another for the xlines, and another for the time samples. Then we can construct the DataArray like so:
import xarray as xr
il, xl, ts = seismic.shape # seismic is a NumPy array.
inlines = np.arange(il) + 1000
xlines = np.arange(xl) + 1000
time = np.arange(ts) * 0.004
da = xr.DataArray(seismic,
name='amplitude',
coords=[inlines, xlines, time],
dims=['inline', 'xline', 'twt'],
)
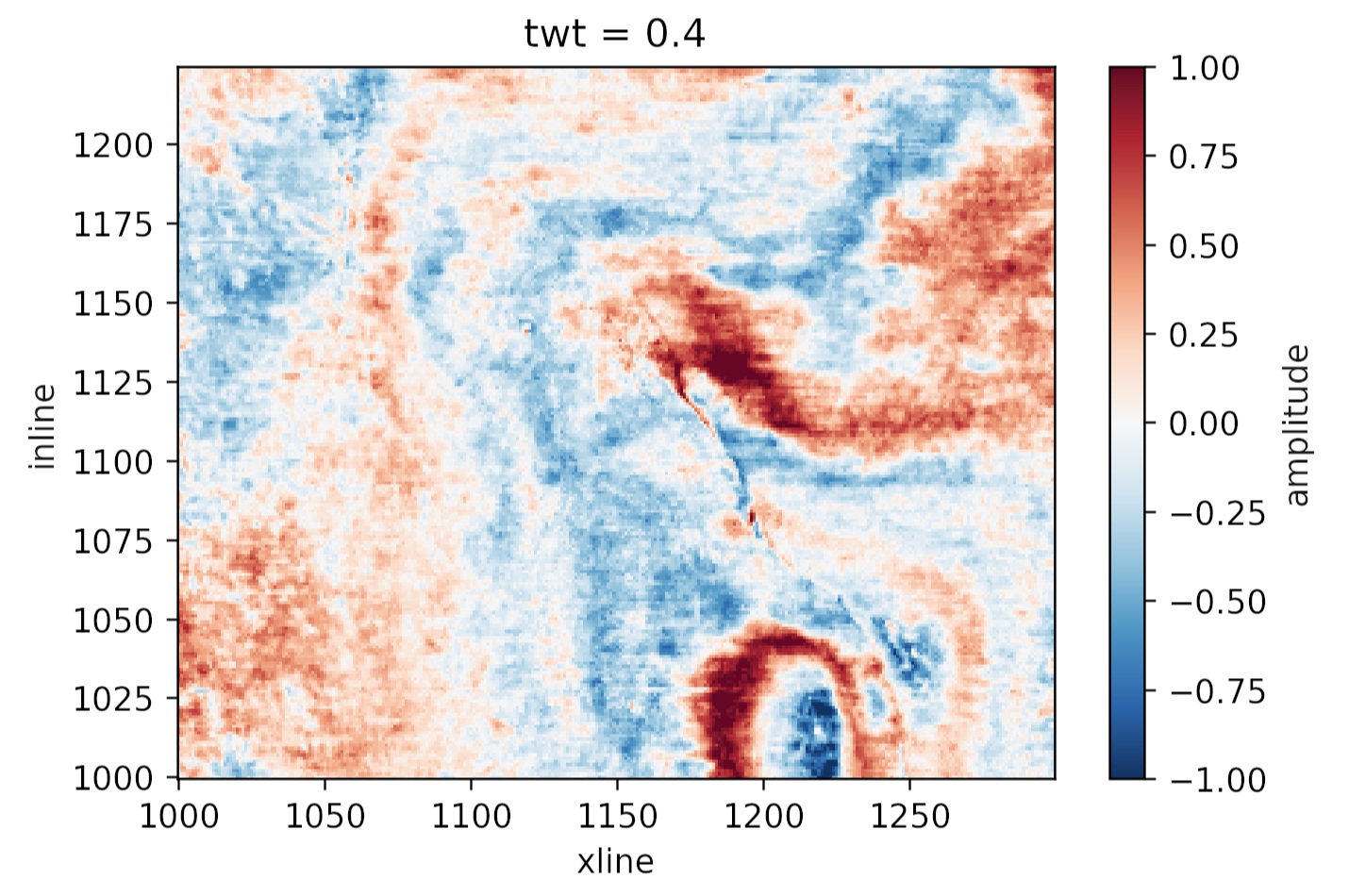
This object da has some nice superpowers. For example, we can visualize the timeslice at 400 ms with a single line:
da.sel(twt=0.400).plot()
to produce this plot:
Plotting the same thing from the NumPy array would require us to figure out which index corresponds to 400 ms, and then write at least 8 lines of matplotlib code. Nice!
What else?
Why else might we prefer this data structure over a simple NumPy array? Here are some reasons:
Extracting amplitude (or whatever) on a horizon. In particular, if the horizon is another
DataArraywith the samedimsas the volume, this is a piece of cake.Extracting amplitude along a wellbore (again, use the same
dimsfor the well path).Making an arbitrary line (a view not aligned with the rows/columns) is easier, because interpolation is ‘free’ with
xarray.Exchanging data with Pandas
DataFrameobjects is easy, making reading from certain formats (e.g. OpendTect horizon export, which gives you inline, xline coords, not a grid) much easier. Even better: use our librarygiofor this! It makesxarrayobjects for you.If you have multiple attributes, then
xarray.Datasetis a nice data structure to keep things straight. You can think of it as a dictionary ofDataArrayobjects, but it’s a lot more than that.You can store arbitrary metadata in
xarrayobjects, so it’s easy to attach things you’d like to retain like data rights and permissions, seismic attribute parameters, data acquisition details, and so on.Adding more dimensions is easy, e.g. offset or frequency, because you can just add more
dimsand associatedcoords. It can get hard to keep these straight in NumPy (was time on the 3rd or 4th axis?).Taking advantage of Dask, which gives you access to parallel processing and out-of-core operations.
All in all, definitely worth a look for anyone who uses NumPy arrays to represent data with real-world coordinates.
Do you already use xarray? What kind of data does it help you with? What are your reasons? Did I miss anything from my list? Let us know in the comments!























 Except where noted, this content is licensed
Except where noted, this content is licensed