x lines of Python: Stereonets
/Difficulty rating: Intermediate
A few years back I needed to plot some fracture data without specialist software, so I created an Excel spreadsheet with a polar plot and interactive widgets. But thanks to Joe Kington and his awesome mplstereonet library those days are over. Today I want to share with you how to plot two fracture sets on an equal area Schmidt plot with mplstereonet.
Here's what we're going to do — and in only 10 lines of Python:
- Load the data from a CSV file.
- Create a stereonet with grid lines.
- Loop over fracture sets and plot each in a different colour.
- Add some statistics for each set.
For data we'll use Irene Wallis's fantastic open-source project fractoolbox repo, which includes some data — as well as some notebooks that go beyond what we will do here.
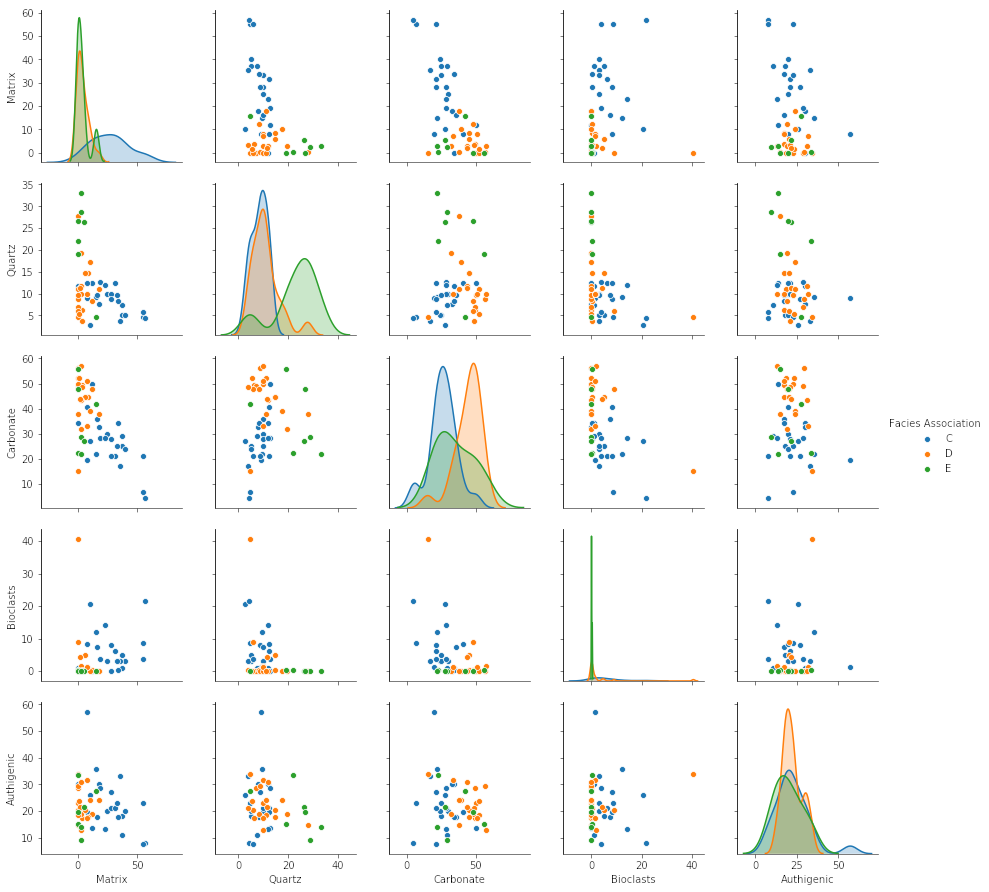
This results in the plot shown here, where each fracture is plotted as a point representing the pole of the fracture plane.
We see that not counting the imports, we can make this simple plot with as a few as 10 lines of code while still retaining some flexibility to refactor this code. The accompanying notebook also shows how to use ipywidgets to make the plot interactive.
That’s it! There’s more in the Notebook — check out the links below. If you get some beautiful plots out of your data, share them in the Software Underground or on Twitter. Have fun!




























 Except where noted, this content is licensed
Except where noted, this content is licensed